Identification of pathogenic bacteria in meats: A Literature review and bibliometric analysis.
Identificación de bacterias patógenas en carnes: Una Revisión de literatura y análisis bibliométrico

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Copyright statement
The authors exclusively assign to the Universidad EIA, with the power to assign to third parties, all the exploitation rights that derive from the works that are accepted for publication in the Revista EIA, as well as in any product derived from it and, in in particular, those of reproduction, distribution, public communication (including interactive making available) and transformation (including adaptation, modification and, where appropriate, translation), for all types of exploitation (by way of example and not limitation : in paper, electronic, online, computer or audiovisual format, as well as in any other format, even for promotional or advertising purposes and / or for the production of derivative products), for a worldwide territorial scope and for the entire duration of the rights provided for in the current published text of the Intellectual Property Law. This assignment will be made by the authors without the right to any type of remuneration or compensation.
Consequently, the author may not publish or disseminate the works that are selected for publication in the Revista EIA, neither totally nor partially, nor authorize their publication to third parties, without the prior express authorization, requested and granted in writing, from the Univeridad EIA.
Show authors biography
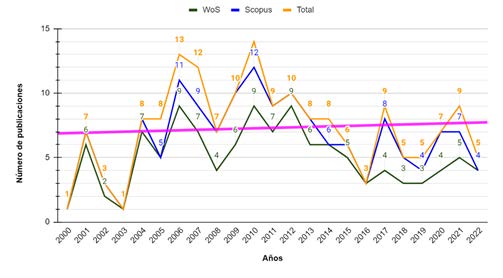
Introduction: Food poisoning caused by pathogenic bacteria present in meats is a growing public health problem. The main method for the identification of these pathogens is the polymerase chain reaction (PCR). Purpose: The purpose of this article is to review the bibliometrics of PCR as applied to the identification of pathogenic bacteria in meats. Methodology: Keywords are identified and search equation is structured in Scopus and WoS databases. Graph analysis was performed using the Bibliometrix, Sci2 Tool and Gephi tools, which are integrated in the R studio software, after which the tree of science metaphor was used.. Results: In the last 10 years, scientific production in areas focused on molecular biology has increased. The most outstanding topics are related to: contaminants and interferences in PCR, the importance of having internal amplification control sequences
in PCR, as well as advances in the standardization of real-time PCR protocols. Salmonella and Listeria monocytogenes stand out as the most investigated in meat matrices. The most used genes in the detection of Salmonella sp are staA, viaB and the sopE for species; for L. monocytogenes it is the hlyA gene. Conclusions: Some of the countries with the highest annual per capita meat consumption are the United States of America, Kuwait, Mexico, Argentina, Austria and Mongolia; however, only the United States of America ranks third in scientific productivity on the subject.
Article visits 364 | PDF visits 133
Downloads
- Acevedo, J., Robledo, S. and Sepúlveda Angarita, M. Z. (2020) “Subáreas de internacionalización de emprendimientos: una revisión bibliográfica,” Economicas Cuc ISSN. Economicas Cuc, 42(1 (2021)), pp. 1–19. doi: 10.17981/econcuc.42.1.2021.Org.7.
- Aras, Z., Sanioğlu Gölen, G. and Kevenk, T. O. (2022) “Salmonella detection in different types of packed raw poultry meat by culture elisa and pcr methods.” Parlar Scientific Publications. Available at: https://hdl.handle.net/20.500.12451/9250 (Accessed: April 7, 2023).
- Aria, M. and Cuccurullo, C. (2017) “bibliometrix : An R-tool for comprehensive science mapping analysis,” Journal of informetrics. Elsevier BV, 11(4), pp. 959–975. doi: 10.1016/j.joi.2017.08.007.
- Bar-Ilan, J. (2010) “Citations to the ‘Introduction to informetrics’ indexed by WOS, Scopus and Google Scholar,” Scientometrics. Springer Science and Business Media LLC, 82(3), pp. 495–506. doi: 10.1007/s11192-010-0185-9.
- Barrera Rubaceti, N. A., Robledo Giraldo, S. and Zarela Sepulveda, M. (2021) “Una revisión bibliográfica del Fintech y sus principales subáreas de estudio,” Económicas CUC. Corporation Universidad de la Costa, CUC, 43(1), pp. 83–100. doi: 10.17981/econcuc.43.1.2022.econ.4.
- Bastian, M., Heymann, S. and Jacomy, M. (2009) “Gephi: An Open Source Software for Exploring and Manipulating Networks,” Proceedings of the International AAAI Conference on Web and Social Media, 3(1), pp. 361–362. doi: 10.1609/icwsm.v3i1.13937.
- Blondel, V. D. et al. (2008) “Fast unfolding of communities in large networks,” Journal of statistical mechanics . IOP Publishing, 2008(10), p. P10008. doi: 10.1088/1742-5468/2008/10/P10008.
- Buitrago, S., Duque, P. L. and Robledo, S. (2019) “Branding Corporativo: una revisión bibliográfica,” Económicas CUC. Corporation Universidad de la Costa, CUC, 41(1). doi: 10.17981/econcuc.41.1.2020.org.1.
- Bundidamorn, D., Supawasit, W. and Trevanich, S. (2018) “A new single-tube platform of melting temperature curve analysis based on multiplex real-time PCR using EvaGreen for simultaneous screening detection of Shiga toxin-producing Escherichia coli, Salmonella spp. and Listeria monocytogenes in food,” Food control. Elsevier BV, 94, pp. 195–204. doi: 10.1016/j.foodcont.2018.07.001.
- Calero Medina, C. M. and Leeuwen, T. N. (2012) “Seed journal citation network maps: A method based on network theory,” Journal of the American Society for Information Science and Technology . Wiley, 63(6), pp. 1226–1234. doi: 10.1002/asi.22631.
- Chandrakar, C. et al. (2022) “ERIC-PCR-based molecular typing of multidrug-resistant Escherichia coli isolated from houseflies (Musca domestica) in the environment of milk and meat shops,” Letters in applied microbiology, 75(6), pp. 1549–1558. doi: 10.1111/lam.13821.
- Clavijo-Tapia, F. J. et al. (2021) “Organizational communication: a bibliometric analysis from 2005 to 2020,” Clío América. Universidad del Magdalena, 15(29), pp. 621–640. doi: 10.21676/23897848.4311.
- Corrales-Reyes, I. E. (2017) “Co-authorship and scientific collaboration networks in Medwave,” Medwave, 17(9), p. e7103. doi: 10.5867/medwave.2017.09.7103.
- Croci, L. et al. (2004) “Comparison of PCR, electrochemical enzyme-linked immunosorbent assays, and the standard culture method for detecting salmonella in meat products,” Applied and environmental microbiology, 70(3), pp. 1393–1396. doi: 10.1128/AEM.70.3.1393-1396.2004.
- Cuadrado Cano, B. S. and Vélez Castro, M. T. (2018) “Contaminación microbiana en la industria de los alimentos.” Corporación Universitaria del Caribe - CECAR, pp. 81–119. Available at: https://repositorio.cecar.edu.co/handle/cecar/2807 (Accessed: June 8, 2023).
- Di Vaio, A. et al. (2021) “The role of digital innovation in knowledge management systems: A systematic literature review,” Journal of business research. Elsevier BV, 123, pp. 220–231. doi: 10.1016/j.jbusres.2020.09.042.
- Dodino-Gutiérrez, C. A. et al. (2023) “Application of molecular techniques in soil microbiology for the identification of bacteria with agricultural potential: a review and bibliometric analysis,” Revista colombiana de ciencias hortícolas, 17(2), pp. e16096–e16096. doi: 10.17584/rcch.2023v17i2.16096.
- Duque, P., Meza, O. E., et al. (2021) “Economía Social y Economía Solidaria: un análisis bibliométrico y revisión de literatura,” REVESCO Revista de Estudios Cooperativos. Universidad Complutense de Madrid (UCM), 138, p. e75566. doi: 10.5209/reve.75566.
- Duque, P., Trejos, D., et al. (2021) “Finanzas corporativas y sostenibilidad: un análisis bibliométrico e identificación de tendencias,” Semestre economico. Universidad de Medellin, 24(56), pp. 25–51. doi: 10.22395/seec.v24n56a1.
- Duque, P. and Cervantes-Cervantes, L.-S. (2019) “Responsabilidad Social Universitaria: una revisión sistemática y análisis bibliométrico,” Estudios gerenciales. Universidad Icesi, pp. 451–464. doi: 10.18046/j.estger.2019.153.3389.
- Duque-Hurtado, P., Toro-Cardona, A., et al. (2020) “Marketing viral: Aplicación y tendencias,” Clío América. Universidad del Magdalena, 14(27), pp. 454–468. doi: 10.21676/23897848.3759.
- Duque-Hurtado, P., Samboni-Rodriguez, V., et al. (2020) “Neuromarketing: Its current status and research perspectives,” Estudios gerenciales. Universidad Icesi, pp. 525–539. doi: 10.18046/j.estger.2020.157.3890.
- Echchakoui, S. (2020) “Why and how to merge Scopus and Web of Science during bibliometric analysis: the case of sales force literature from 1912 to 2019,” Journal of Marketing Analytics. Palgrave, 8(3), pp. 165–184. doi: 10.1057/s41270-020-00081-9.
- Fajardo Rico, A. (2019) “Estudio de factibilidad financiera para la creación de una empresa empacadora al vacío de carne bovina madurada en florencia, caquetá.” Universidad Nacional Abierta y a Distancia UNAD. Available at: https://repository.unad.edu.co/handle/10596/28062 (Accessed: August 22, 2023).
- Fan, W. et al. (2022) “Rapid and simultaneous detection of Salmonella spp., Escherichia coli O157:H7, and Listeria monocytogenes in meat using multiplex immunomagnetic separation and multiplex real-time PCR,” European food research and technology = Zeitschrift fur Lebensmittel-Untersuchung und -Forschung. A. Springer Science and Business Media LLC, 248(3), pp. 869–879. doi: 10.1007/s00217-021-03933-5.
- FAO (2019) Cambio climático y seguridad alimentaria y nutricional en América Latina y el Caribe, Organización de las Naciones Unidas para la Alimentación y la Agricultura. Available at: https://www.fao.org/documents/card/es?details=CA2902ES/ (Accessed: February 13, 2023).
- Freeman, L. C. (1977) “A set of measures of centrality based on betweenness,” Sociometry. JSTOR, 40(1), p. 35. doi: 10.2307/3033543.
- González Salas, R., Vidal del Río, M. M. and Monsalve Guamán, A. A. (2023) “Diagnóstico y transmisión de la infección alimentaria salmonelosis,” Dilemas contemporáneos: educación, política y valores. Asesorias y tutorias para la Investigacion Cientifica en la Educacion Puig-Salabarria S.C. doi: 10.46377/dilemas.v2i10.3564.
- González-Valiente, C. L. (2019) “Redes de citación de revistas iberoamericanas de Bibliotecología y Ciencia de la Información en Scopus,” Bibliotecas. Anales de investigación, 15(1), pp. 83–98. Available at: http://revistas.bnjm.cu/index.php/BAI/article/view/115 (Accessed: April 9, 2023).
- Gurzki, H. and Woisetschläger, D. M. (2017) “Mapping the luxury research landscape: A bibliometric citation analysis,” Journal of business research. Elsevier BV, 77, pp. 147–166. doi: 10.1016/j.jbusres.2016.11.009.
- Hirsch, J. E. (2005) “An index to quantify an individual’s scientific research output,” Proceedings of the National Academy of Sciences of the United States of America, 102(46), pp. 16569–16572. doi: 10.1073/pnas.0507655102.
- Hoorfar, J. et al. (2003) “Making internal amplification control mandatory for diagnostic PCR,” Journal of clinical microbiology, 41(12), p. 5835. doi: 10.1128/JCM.41.12.5835.2003.
- Hoorfar, J. et al. (2004) “Practical considerations in design of internal amplification controls for diagnostic PCR assays,” Journal of clinical microbiology, 42(5), pp. 1863–1868. doi: 10.1128/JCM.42.5.1863-1868.2004.
- Hoorfar, J., Ahrens, P. and Rådström, P. (2000) “Automated 5’ nuclease PCR assay for identification of Salmonella enterica,” Journal of clinical microbiology, 38(9), pp. 3429–3435. doi: 10.1128/JCM.38.9.3429-3435.2000.
- Irkin, R., Bozkurt, B. and Tumen, G. (2021) “Determination of the prevalence of Salmonella spp. and S. aureus in meat products by Real-Time PCR and testing their antibiotic susceptibility,” Medycyna weterynaryjna. Medycyna Weterynaryjna - Redakcja, 77(06), pp. 6533–2021. doi: 10.21521/mw.6533.
- Jaroenporn, C. et al. (2022) “In-House Validation of Multiplex PCR for Simultaneous Detection of Shiga Toxin-Producing , and spp. in Raw Meats,” Foods (Basel, Switzerland), 11(11). doi: 10.3390/foods11111557.
- Khan, A. S. (2017) “Biotechnology-based sensing platforms for detecting foodborne pathogens,” in Analysis of Food Toxins and Toxicants. Chichester, UK: John Wiley & Sons, Ltd, pp. 37–50. doi: 10.1002/9781118992685.ch2.
- Labrador, M. et al. (2018) “Comparative evaluation of impedanciometry combined with chromogenic agars or RNA hybridization and real-time PCR methods for the detection of L. monocytogenes in dry-cured ham,” Food control. Elsevier BV, 94, pp. 108–115. doi: 10.1016/j.foodcont.2018.06.031.
- Labrador, M., Giménez-Rota, C. and Rota, C. (2021) “Real-Time PCR Method Combined with a Matrix Lysis Procedure for the Quantification of in Meat Products,” Foods (Basel, Switzerland), 10(4). doi: 10.3390/foods10040735.
- Landinez, D. A., Robledo Giraldo, S. and Montoya Londoño, D. M. (2019) “Executive Function performance in patients with obesity: A systematic review,” Psychologia. Universidad de San Buenaventura, 13(2), pp. 121–134. doi: 10.21500/19002386.4230.
- Lazou, T. P. et al. (2021) “Method-Dependent Implications in Foodborne Pathogen Quantification: The Case of Survival on Meat as Comparatively Assessed by Colony Count and Viability PCR,” Frontiers in microbiology, 12, p. 604933. doi: 10.3389/fmicb.2021.604933.
- Lopes, A. T. S., Albuquerque, G. R. and Maciel, B. M. (2018) “Multiplex Real-Time Polymerase Chain Reaction for Simultaneous Quantification of spp., , and in Different Food Matrices: Advantages and Disadvantages,” BioMed research international, 2018, p. 6104015. doi: 10.1155/2018/6104015.
- López, A. et al. (2018) “Contaminación microbiológica de la carne de pollo en 43 supermercados de El Salvador,” ALERTA Revista Científica del Instituto Nacional de Salud. Latin America Journals Online, 1(2), pp. 45–53. doi: 10.5377/alerta.v1i2.7134.
- Malorny, B. et al. (2003) “Standardization of diagnostic PCR for the detection of foodborne pathogens,” International journal of food microbiology, 83(1), pp. 39–48. doi: 10.1016/s0168-1605(02)00322-7.
- Malorny, B. et al. (2004) “Diagnostic real-time PCR for detection of Salmonella in food,” Applied and environmental microbiology, 70(12), pp. 7046–7052. doi: 10.1128/AEM.70.12.7046-7052.2004.
- Miguel, S., Moya-Anegón, F. and Herrero-Solana, V. (2007) “El análisis de co-citas como método de investigación en Bibliotecología y Ciencia de la Información,” Investigación bibliotecológica. Universidad Nacional Autónoma de México, Instituto de Investigaciones Bibliotecológicas y de la Información, 21(43), pp. 139–155. Available at: http://www.scielo.org.mx/scielo.php?script=sci_abstract&pid=S0187-358X2007000200006&lng=es&nrm=iso&tlng=es (Accessed: March 4, 2023).
- Öz, Y. Y. et al. (2020) “Rapid and sensitive detection of Salmonella spp. in raw minced meat samples using droplet digital PCR,” European food research and technology = Zeitschrift fur Lebensmittel-Untersuchung und -Forschung. A. Springer Science and Business Media LLC, 246(10), pp. 1895–1907. doi: 10.1007/s00217-020-03531-x.
- Perelle, S. et al. (2004) “Comparison of PCR-ELISA and LightCycler real-time PCR assays for detecting Salmonella spp. in milk and meat samples,” Molecular and cellular probes, 18(6), pp. 409–420. doi: 10.1016/j.mcp.2004.07.001.
- Petsios, S. et al. (2016) “Conventional and molecular methods used in the detection and subtyping of Yersinia enterocolitica in food,” International journal of food microbiology, 237, pp. 55–72. doi: 10.1016/j.ijfoodmicro.2016.08.015.
- Queiroz, M. M. and Fosso Wamba, S. (2021) “A structured literature review on the interplay between emerging technologies and COVID-19 - insights and directions to operations fields,” Annals of Operations Research, pp. 1–27. doi: 10.1007/s10479-021-04107-y.
- Rabelo Florez, R. A. (2022) “Bacterias y hongos utilizados en la biodegradación de hidrocarburos: Una Revisión de literatura y Análisis Bibliométrico,” Revista EIA. Universidad EIA, 20(39). doi: 10.24050/reia.v20i39.1622.
- Rahn, K. et al. (1992) “Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella,” Molecular and cellular probes, 6(4), pp. 271–279. doi: 10.1016/0890-8508(92)90002-f.
- Ramos-Enríquez, V., Duque, P. and Vieira Salazar, J. A. (2021) “Responsabilidad Social Corporativa y Emprendimiento: evolución y tendencias de investigación,” Desarrollo gerencial. Universidad Simon Bolivar, 13(1), pp. 1–34. doi: 10.17081/dege.13.1.4210.
- Robledo, S., Osorio, G. and Lopez, C. (2014) “Centro de Investigación de la Universidad Distrital Francisco José de Caldas,” Revista Vínculos, 11(2), pp. 6–16. doi: 10.14483/2322939X.9664.
- Rodríguez Sánchez, I. P. and Barrera Saldaña, H. A. (2004) “La reacción en cadena de la polimerasa a dos décadas de su invención,” Ciencia UANL, 7(3). Available at: http://eprints.uanl.mx/1584/1/art_cadena.pdf (Accessed: February 13, 2023).
- Rossen, L. et al. (1992) “Inhibition of PCR by components of food samples, microbial diagnostic assays and DNA-extraction solutions,” International journal of food microbiology, 17(1), pp. 37–45. doi: 10.1016/0168-1605(92)90017-w.
- Secinaro, S. et al. (2022) “Blockchain in the accounting, auditing and accountability fields: a bibliometric and coding analysis,” Accounting auditing & accountability. Emerald, 35(9), pp. 168–203. doi: 10.1108/aaaj-10-2020-4987.
- Soumet, C. et al. (1999) “Evaluation of a multiplex PCR assay for simultaneous identification of Salmonella sp., Salmonella enteritidis and Salmonella typhimurium from environmental swabs of poultry houses,” Letters in applied microbiology, 28(2), pp. 113–117. doi: 10.1046/j.1365-2672.1999.00488.x.
- Stingl, K. et al. (2021) “Challenging the ‘gold standard’ of colony-forming units - Validation of a multiplex real-time PCR for quantification of viable Campylobacter spp. in meat rinses,” International journal of food microbiology, 359, p. 109417. doi: 10.1016/j.ijfoodmicro.2021.109417.
- Sustacha, I. et al. (2022) “Research trends in technology in the context of smart destinations: a bibliometric analysis and network visualization,” Cuadernos de gestión. Enpresa Institutua - Instituto de Economia Aplicada a la Empresa, 22(1), pp. 161–173. doi: 10.5295/cdg.211501is.
- Syarifah, I. K. et al. (2020) “Identification and differentiation of isolated from chicken meat using real-time polymerase chain reaction and high resolution melting analysis of and genes,” Veterinary world, 13(9), pp. 1875–1883. doi: 10.14202/vetworld.2020.1875-1883.
- Tani, M., Papaluca, O. and Sasso, P. (2018) “The System Thinking Perspective in the Open-Innovation research: A systematic review,” Journal of Open Innovation Technology Market and Complexity. Elsevier BV, 4(3), p. 38. doi: 10.3390/joitmc4030038.
- Tobing, E. C. L. and Arianto, I. D. (2022) “Analisis Jaringan Komunikasi German Digital #PERCUMALAPORPOLISI di Twitter,” Jurnal Nomosleca, 8(2), pp. 146–159. doi: 10.26905/nomosleca.v8i2.7677.
- Torres, G., Robledo, S. and Rojas Berrío, S. (2022) “Market orientation: importance, evolution, and emerging approaches using scientometric analysis,” Criterio libre. Universidad Libre, 19(35), pp. 326–340. doi: 10.18041/1900-0642/criteriolibre.2021v19n35.8371.
- Trejos-Salazar, D. F. et al. (2021) “Neuroeconomía: una revisión basada en técnicas de mapeo científico,” Revista de Investigación Desarrollo e Innovación. Universidad Pedagogica y Tecnologica de Colombia, 11(2), pp. 243–260. doi: 10.19053/20278306.v11.n2.2021.12754.
- Valencia-Hernandez, D. S. et al. (2020) “SAP algorithm for citation analysis: An improvement to tree of Science,” Ingeniería e Investigación. Universidad Nacional de Colombia, 40(1). doi: 10.15446/ing.investig.v40n1.77718.
- Vera-Baceta, M.-A., Thelwall, M. and Kousha, K. (2019) “Web of Science and Scopus language coverage,” Scientometrics. Springer Science and Business Media LLC, 121(3), pp. 1803–1813. doi: 10.1007/s11192-019-03264-z.
- Wallis, W. D. (2007) A beginner’s guide to graph theory. 2nd ed. Secaucus, NJ: Birkhauser Boston. doi: 10.1007/978-0-8176-4580-9.
- Whyte, P. et al. (2002) “The prevalence and PCR detection of Salmonella contamination in raw poultry,” Veterinary microbiology, 89(1), pp. 53–60. doi: 10.1016/s0378-1135(02)00160-8.
- Wilson, I. G. (1997) “Inhibition and facilitation of nucleic acid amplification,” Applied and environmental microbiology, 63(10), pp. 3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997.
- Yang, S., Zheng, L. and Keller, F. B. (2016) Social Network Analysis: Methods and Examples. Sage Publications, Incorporated. Available at: https://books.google.com/books/about/Social_Network_Analysis.html?hl=&id=2PUHogEACAAJ.
- Zariñana, R. and De la, A. E. (2017) Estandarización y validación de la técnica de PCR para la determinación de Listeria monocytogenes en carne de pollo, res y cerdo. Available at: http://hdl.handle.net/10521/3846 (Accessed: February 13, 2023).
- Zhanabayeva, D. K. et al. (2021) “PCR Diagnosis for the Identification of the Virulent Gene of Salmonella in Poultry Meat.” OnLine Journal of Biological Sciences. Available at: http://repository.kazatu.kz/jspui/handle/123456789/1535 (Accessed: April 7, 2023).
- Zhang, J. and Luo, Y. (2017) “Degree centrality, betweenness centrality, and closeness centrality in social network,” in Proceedings of the 2017 2nd International Conference on Modelling, Simulation and Applied Mathematics (MSAM2017). 2017 2nd International Conference on Modelling, Simulation and Applied Mathematics (MSAM2017), Paris, France: Atlantis Press. doi: 10.2991/msam-17.2017.68.
- Zhu, J. and Liu, W. (2020) “A tale of two databases: the use of Web of Science and Scopus in academic papers,” Scientometrics. Springer Science and Business Media LLC, 123(1), pp. 321–335. doi: 10.1007/s11192-020-03387-8.
- Zupic, I. and Čater, T. (2015) “Bibliometric methods in management and organization,” Organizational research methods. SAGE Publications, 18(3), pp. 429–472. doi: 10.1177/1094428114562629.
- Zuschke, N. (2020) “An analysis of process-tracing research on consumer decision-making,” Journal of business research. Elsevier BV, 111, pp. 305–320. doi: 10.1016/j.jbusres.2019.01.028.




